17-04-2023
Latvian Biomedical Research and Study Center begins involvement of participants in the Latvian Population Genome Reference Project, which takes place within the framework of the European ‘1+ Million Genomes’ initiative

The main goal of the project is to create a genome reference of the Latvian population that will be used for disease prognostication, screening planning, treatment optimization, and scientific development. The Latvian genome reference created as part of the project, together with the population genome data of other European countries, will be included in the unified European genome database within the framework of the '1+ Million Genomes' (1+MG) initiative. The goal of the 1+MG initiative is to provide secure access to genetic and sample-related clinical data across Europe to improve research, develop personalized healthcare, and support more effective decision-making in healthcare policy (more information on the 1+MG initiative: here
More information: here
29-04-2021
To date, 16 different strains of Covid-19 virus have been registered in Latvia. There are currently several thousand mutations of Covid-19 virus around the world, but in order for a common mutation to be considered as a new type of virus, it is evaluated against a number of criteria. As the Vice-Chair of the Latvian Biomedical Research and Study Center Dr. Jānis Kloviņš reveals, one of the criteria is the prevalence of the mutant variant. To date, 16 different strains of Covid-19 virus have been registered in Latvia. You can find more information here.
28-04-2021
According to data provided to the government of the Latvian Biomedical Research and Study Center, the British Covid-19 variant reaches almost 70% of the analyzed material.The proportion of isolates (materials) corresponding to the British type continues to grow. So far, 700 isolates have been detected. In the previous week, there were 63, which is about 68% of the analyzed samples. More information can be found here.
12-04-2021
The project Izp-2018/2-0171 ''Ethically and socially responsible management of research biobanks in Latvia: analysis of public, donor and scientist opinions''has been completed.
During the research, number of the following public reports has been created:
Report on Biobank donor opinion - https://genomadatubaze.lv/uploads/materials/Zinojums-par-biobanku-donoru-viedokli-FINAL-1.pdf
Report on public survey - https://genomadatubaze.lv/uploads/materials/Zinojums-par-sabiedribas-aptauju-FINAL.pdf
Report of Scientist opinion - https://genomadatubaze.lv/uploads/materials/Zinojums-par-zinatnieku-viedokli-FINAL.pdf
Recomendations for Biobank management - https://genomadatubaze.lv/uploads/materials/Rekomendacijas_biobanku-etiska-parvaldiba.pdf
23-03-2021
First record in Latvia, when the Covid-19 vaccine did not work due to a mutation of the virus.
Latvian Television News service has led an interview with members of Latvian Biomedicine research and study center:
- Chairman of the Scientiffic Council Prof. Jānis Kloviņš
- Researcher Monta Ustinova
- Research Assistant Nikita Zrelov.
The record can be found here.
17-03-2021
New cases of Covid-19 Uganda virus A.23.1 has been found at laboraroty of Latvian Biomedicine and research center. Cases are local to Liepāja city.
Two weeks ago, during evaluation of sample test results, Covid-19 Uganda virus A.23.1 has been identified in three samples of 192 (1,56%) while this week Uganda virus has been found in four cases of 96 (4%). All cases are registered from Liepāja city. Information is provided by Chairman of Scientiffic council Jānis Kloviņš.
Such Covid-19 type originally was recorded from Uganda, but it differs from Southafrican Covid-19 type, that is not found in Latvia.
Uganda Covid-19 type is widespread at United Kingdom, and by unknown reasons has been distributed to Latvia.
19-02-2021
This week, trought analyses of 96 Covid-19 samples, were found three new cases of Great Britan Covid-19 type. Information is provided by Latvian Biomedicine research and Study centre Scientiffic board chairman Jānis Kloviņš.
So far, a total of 12 cases of British Covid-19 infection have been found in Latvia.
Center of Disease control and prevention (SPKC) has previously emphasized that the new variant of Covid-19 found in UK does not differ in severity from the current one, but has a higher contagion, hence, the new Covid-19 strain is no more dangerous than the existing one.

More information can be found here.
02-07-2020
Genome Database has started implementation of National Research Programm "For mitigation of COVID-19".
The goal of the project is to establish an open biobank and data exchange resource to facilitate the research on COVID -19, create a framework for its integration with health care initiatives, and ensure international cooperativity.
Principal investigator: Dr. biol. Jānis Kloviņš
Research is funded by National Research Programm No. VPP-COVID-2020/1-0016
Approval by Central Medical Ethics Committee No: 01-29.1/5034
16-01-2020
Results of a public survey about research biobanks are available.
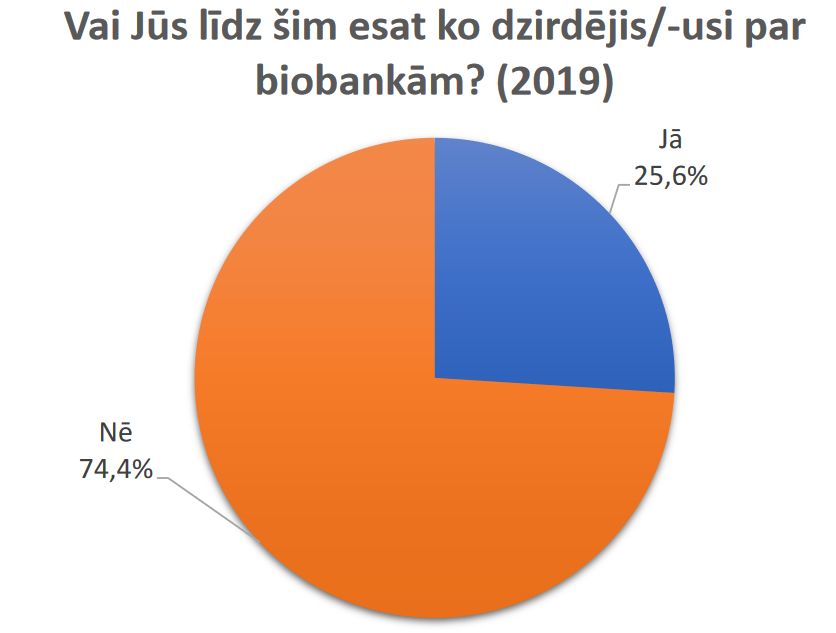
Latvian Biomedical research and study centre and Institute of Clinical and Preventive Medicine, University of Latvia are implementing project “Ethically and socially responsible governance of research biobanks in Latvia: analysis of opinions of public, donors and researchers” (funded by Latvian Council of Science No. 2018/2-0171). In the framework of this project in May 2019 the survey on public attitudes towards biobanks in Latvia was carried out.
Results of the survey are available here (in Latvian).
Information about the project here.


01-01-2020
On the 1st of January, 2020, Latvian Biomedical Research and Study centre has started research project "Multidisciplinary study of community-acquired sepsis survivors in Latvia" (Funded by Grant from Latvian Council of Science: lzp-2019/1-0225). The leading partner of the project is University of Latvia, head of the project Dr. med. Uga Dumpis, scientific leader of the project from Biomedical centre side is Dr. biol., Prof. Jānis Kloviņš.
This research will explore impacts, needs, and burden of care among sepsis survivors by offering multidisciplinary approach and investigating social, clinical and biological mechanisms causing long-term morbid events. The primary objective of the study is to obtain in-depth understanding on the sepsis survivors, including clinical, molecular aspects of the recovery from illness. Secondary objectives are: 1. Explore the general care pathways and outcomes of sepsis patients using the sepsis register of the PSCUH and National data base. 2. Investigate perceptions, experience and needs of sepsis survivors, their informal and formal caregivers during the post-sepsis recovery. 3. Analyse molecular data using blood and stool samples of sepsis survivors for one year. Exploratory objectives are: 1. Find novel candidate biomarkers for characterization of the course of recovery from sepsis. The research project will not only offer first such study in Latvian context but also provide comprehensive understanding of molecular, clinical, and social pathways of post-sepsis survival.


01-01-2020
On the 1st of January, 2020, Latvian Biomedical Research and Study centre has started research project "Investigation of the origin and dynamics of human blood microbiome, and its role in chronic disease" (Funded by Grant from Latvian Council of Science: lzp-2019/1-0116). The scientific leader of the project is Dr. biol., Prof. Jānis Kloviņš.
Summary
Reports on the existence of a persistent human blood-microbiome even in healthy persons are steadily accumulating. However, the origins, identities, and functions, as well as connection to pathologies still remain to be elucidated. The main aim of this project is to understand the origin and dynamics of human blood microbiome and its association with intestinal inflammation in order to identify reliable omics-based biomarkers for early detection and monitoring of chronic conditions using type 2 diabetes, irritable bowel syndrome and inflammatory bowel disease models. This will be achieved through 1) clarification of the origin of exogenous nucleic acids in blood by obtaining skin, oral and gut microbiome compositions from study participants; 2) investigation of the functional inflammatory landscape of study participants in correspondence to blood microbiome profiles, focusing on intestinal integrity markers 3) investigation of the association of blood microbiome composition with intestinal diseases and T2D as well as in response to anti-diabetic and anti-inflammatory therapy for the potential development of omics-based biomarkers.
As a result, the project will provide not only deeper insight into the origin and stability of blood microbiome, but also on its putative association with gut inflammation and related diseases.


02-09-2019
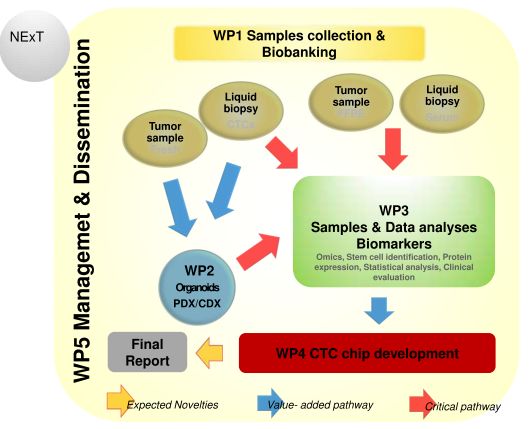
On the 1st of September, 2019, Latvian Biomedical Research and Study centre has started researh project "Establishing an algorithm for the early diagnosis and follow-up of patients with pancreatic neuroendocrine tumors - NExT" (Funded by Joint Transnational Call for Proposals 2017). The scientific groups from Germany, Spain, Slovakia, Latvia and Greece, project coordinator is Fraunhofer-Gesellschaft zur Förderung der angewandten Forschung e.V., Institute for Biomedical Engineering IBMT from Germany. The scientific leader from Latvia is Dr. biol. Vita Rovīte.
Standardized clinical management of pancreatic neuroendocrine tumours (PNETs) is limited by different aspects of the disease, as its relative rarity (1 per 100,000 individuals), heterogeneous clinical presentation (hormonally functional or non-functional), the limited understanding of tumour biology and behaviour and the lack of prospectively evaluated risk stratification systems. While surgical excision remains the primary therapy, as early detection is uncommon, most patients present with metastatic disease at diagnosis and a reduced life expectancy as they are not candidates for resection.
Hypothesis: Prompt, specific and sensitive detection and characterisation of PNETs could lead to early detection, increasing the chance for surgical intervention and improve patients’ survival. Circulating tumour cells (CTCs) shed from primary tumours are considered attractive biomarkers for liquid biopsy as they represent an early step in blood-borne metastasis. Several studies propose that during malignant progression cancer cells undergo an epithelial-to-mesenchymal transition (EMT), acquire invasive properties and stem cell-like features.
Aims: The present proposal, by building up a tissue bank of genetically characterized tumours, development of patient-derived rare tumour xenografts (PDXs) and organoids, aim to identify PNET-specific biomarkers urgently needed to design a next generation nanotechnology based microfluidic device and integrate the technology of minimally invasive liquid biopsy in the early detection of PNETs.
Methods: Serum and tissue samples (fresh and FFPE) will be collected from PNET patients and their clinical and lab records will be recorded. An expression study and genomic analysis will be performed in clinical samples by different molecular techniques whereas in parallel organoids, PDX and CDX will be established. A next generation nanotechnology based microfluidic device will be developed, based on the acquired data.
Expected results and potential impact: Better understanding of the etiopathogenetic determinants involved in PNETs formation resulting from the multidisciplinary collaboration within ‘NExT” consortium will hopefully deliver a nanotechnology-based microfluidic (next generation) device that by means of CTCs detection will present a powerful tool for the early detection of PNET tumours and follow-up of patients, contributing to their better medical treatment.


02-04-2019
On 1st April in Latvian Biomedical Research and Study centre in collaboration with Genome Database of Latvian population will begin new project funded by European Regional Development Fund (ERDF) Operational Programme Growth and Employment 1.1.1 Specific Aid Objective to increase the research and innovative capacity of scientific institutions of Latvia and the ability to attract external financing, investing in human resources and infrastructure Measure 1.1.1.1 “Support for applied research” 2. Selection round.
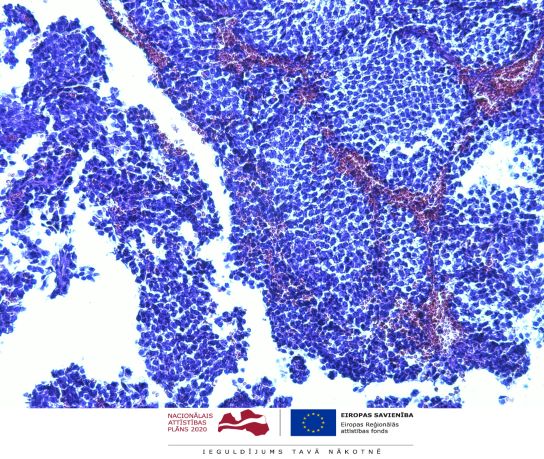
"RNA molecular determinants in development of pituitary adenoma” (Project Identification No 1.1.1.1/18/A/089). The project principle investigator in BMC is Dr. biol. Vita Rovīte. Cooperation partner is SIA “GenEra”.
About the project:
The scientific objective of the project is to study the spectrum of various ribonucleic acid (RNA) markers (mRNA, non-coding) in development of pituitary adenoma (PA) to discover determinants that influence therapy response and disease outcome.
The main activities to achieve the goal of the project: 1) identify and analyse molecular RNA factors in primary PA tissue by massive parallel sequencing (MPS), 2) study RNA types in primary culture of free-floating spheres and mesenchymal cells from PA tissue to describe tumour-borne RNA patterns, 3) investigate alterations in coding and non-coding RNA in free-floating sphere model upon addition of PA treatment medication, 4) validate discovered markers in PA patients’ plasma and independent cohort, 5) functionally characterize discovered RNA candidates in GH3 cells.
The results of the project will evaluate molecular RNA factors on several functional levels of PA derived biological materials and models: (1) potential makers discovered in tissues and validated in plasma and/or tissues could serve for prediction of disease outcome, characterization of aggressiveness and non-invasive diagnostics, (2) study of free-floating spheres and mesenchymal cells will help to estimate spheres as isolated model for PA tumour-borne cells, define their RNA profile and describe PA therapy response in this system, (3) research of influence of discovered RNA candidates in GH3 cells will functionally characterize the precise cellular mechanisms of these factors in PA development.
02-04-2019
On 1st April BMC will be launched European Regional Development Fund (ERDF) Operational Programme Growth and Employment 1.1.1 Specific Aid Objective to increase the research and innovative capacity of scientific institutions of Latvia and the ability to attract external financing, investing in human resources and infrastructure Measure 1.1.1.1 “Support for applied research” 2. Selection round.
"The determination of rare inherited diseases' causative mechanisms using whole genome sequencing approach” (Project Identification No 1.1.1.1/18/A/096). The project principle investigator in BMC is Dr. biol. Inna Iņaškina. Cooperation partner is SIA Latvia MGI Tech.
About the project:
The general aim of the project is to improve public health through the expansion of our knowledge about rare inherited diseases and diagnostics of patients with such disorders. Specific aim of the project is to identify novel causative genetic elements in rare inherited disease patients in whom pathogenic variants have not been identified so far by conventional genetic analysis approaches.
To reach the specific aim of the project we have tailored the following activities:
1. Establishing criteria for selection and recruitment of patients suffering from rare diseases. The patients included in the study will represent a challenging group for testing as they have been previously extensively investigated using gene panel or whole exome sequencing approaches and remain without a genetic diagnosis for their disease.
2. Whole genome sequencing of trio samples (patients plus parents), bioinformatic analysis of data
3. Validation and interpretation of genetic variants identified by WGS. Analysis of candidate genes or other genetic elements selected on the basis of WGS, using available sources of scientific information (publications, genetic variant and medical databases, as well as databases on available animal models).

02-04-2019
On 1st April in Latvian Biomedical Research and Study centre in collaboration with Genome Database of Latvian population will begin project funded by European Regional Development Fund (ERDF) Operational Programme Growth and Employment 1.1.1 Specific Aid Objective to increase the research and innovative capacity of scientific institutions of Latvia and the ability to attract external financing, investing in human resources and infrastructure Measure 1.1.1.1 “Support for applied research” 2. Selection round.

" Role of miRNAs in the host-gut microbiome communication during metformin treatment in the context of metabolic disorders” (Project Identification No 1.1.1.1/18/A/092). The project principle investigator in BMC is Dr. biol. Jānis Kloviņš. Cooperation partner is SIA “GenEra”.
About the project:
Changes in core gut microbiome are associated with a wide variety of diseases, however, the mechanisms of interaction in the ability of the host to regulate these deviations are still unclear and under-investigated. Recently host miRNAs were shown to be involved in the communication between the host and its gut microbiome. There are still many caveats in the understanding of the possible mechanisms and the actual impact of observed association and further research is needed. Equally important is to understand the communication in relation to gut microbiome-related diseases such as type 2 diabetes (T2D) not only to understand the mechanisms of these phenomena in disease, but also to open opportunity for creation of completely novel therapeutic strategies.
The overall aim of the project is to investigate the contribution of the host miRNAs in metformin-mediated changes in gut microbiome composition and functions in the context of type 2 diabetes and to identify miRNAs which could be directed to potential clinical studies.
We will use notable sample resources collected in the framework of a previously performed mice experiment. In addition we will use samples from patients with different metabolic backgrounds obtained during colonoscopy procedure.
We will obtain gut metagenome and miRNA composition data by NGS and estimate the correlation between miRNA and gut microbiome profiles in high-fat diet-fed mice model and patients.
This project will result in a novel understanding of host miRNA-microbiome interaction in T2D and deliver new knowledge for development of personalized approaches in T2D treatment or improvement of the existing strategies.
02-04-2019
On 1st April in Latvian Biomedical Research and Study centre in collaboration with Genome database of Latvian population will begin new project of European Regional Development Fund (ERDF) Operational Programme Growth and Employment 1.1.1 Specific Aid Objective to increase the research and innovative capacity of scientific institutions of Latvia and the ability to attract external financing, investing in human resources and infrastructure Measure 1.1.1.1 “Support for applied research” 2. Selection round.

"Functional and genetic research of rare unidentitied neuromuscular disorders” (Project Identification No 1.1.1.1/18/A/097). The project principle investigator in BMC is Dr. biol. Inna Iņaškina. Cooperation partner is SIA Latvia MGI Tech.
About the project:
The general aim of the project is to improve public health through the expansion of our knowledge about rare neuromuscular diseases and diagnostics of patients with such disorders. The specific aim of the project focuses on two topics: 1) to better understand and characterize the recently identified mutant MYBPC1 protein, its interactions with other cellular partners, and the molecular mechanisms underlying newly described disease phenotype; 2) using the newest molecular biology methods, namely whole genome sequencing, to identify novel causative genetic elements (genes, inter-gene regions or chromosomal rearrangements) for rare and/or unidentified neuromuscular disorders.
To reach the specific aim of the project, we have tailored the following activities: development of knockin mouse model for the mutation in human MYBPC1 gene and its in-depth study; transcriptome analysis of patients’ tissue samples and tissue samples obtained from the mouse models; recruitment of patients suffering from rare and/ or unidentified neuromuscular diseases; whole genome sequencing analysis of aforementioned patients with a goal to identify novel causative genetic elements.
Research will join advances and technologies from several scientific fields: medicine, biomedicine, biotechnology and bioinformatics. Public access to research results will be provided in the form of animal functional model data, publications and patients’ whole genome sequencing data available for further studies (in an anonymized form).

08-01-2019
Genome Database of Latvian population participates in project "Interplay of Environmental and Genetic Factors in the Immunologic Mechanisms of Thyroid Autoimmune Diseases".

Aim of the project is to analyze susceptibility genetic variants for Autoimmune Thyroid Diseases (AITD) in patients with Hashimoto’s thyroiditis (HT) and Graves’ disease (GD) according to precision medicine concept and to investigate the interaction between them and particular environmental factors including selenium intake and status, stress, lifestyle habits and other triggers in the occurrence of different T helper pathways and cytokine patterns.
Objectives to reeach the aim are:
1. To investigate association of immune-modulating and selenoprotein genotypes with AITD and related clinical, environmental parameters and T helper pathways.
2. To detect whether iodine supplementation improves iodine status in pregnant women with mild iodine deficiency and whether iodine used in combination with selenium will reduce risk of thyroid autoimmunity during pregnancy or postpartum period in women with assessed selenium status and genetic variants.
3. To investigate the long-term effect of selenium supplementation on the thyroid function and disease activity as well as to evaluate its modulating effects on shifting T helper immune response in treatment-naïve euthyroid patients with HT in context with selenium status and assessed immune-modulating and selenoprotein genetic variants.
4. To evaluate the possible association between the number and impact of stressful life events in AITD patients and Th1, Th2, and Th17 circulating cytokine profiles in order to better understand the effects of stress-related mechanisms on the immune system in association with cytokine-related genes.
Principle Investigator: Assoc. prof., Dr. med. Ilze Konrāde
Leading partner of the project is Riga Stradins University.
Project is funded by Latvian Council of Science grant no. lzp-2018/2-0059.


03-12-2018
On December, 2018 Latvian Biomedical Research and Study centre has started the implementation of the Latvian Council of Science funded project “Ethically and socially responsible governance of research biobanks in Latvia: analysis of opinions of public, donors and researchers” (lzp-2018/2-0171). The leading partner of the project is Institute of Clinical and Preventive Medicine of University of Latvia, from the Latvian Biomedical Research and Study centre the implementation is carried out by Genome Database of Latvian population.
Nowadays research biobanks are crucial for high level research in areas of biomedicine and public health. The overall aim of the project is to contribute to ethically and socially responsible governance of research biobanks in Latvia by analysing attitudes, concerns, and needs of the general public, donors and researchers. The aim will be obtained by the study of the publics’, donors’ and researchers’ attitudes, concerns and trust in biobanking and analysis will be performed how the concerns and needs raised by the public, donors and researchers can be adequately dealt by the normative framework to finally improve research biobank governance and drive public policy decisions by developing written recommendations based on the results of the study. The study will be performed by interdisciplinary team that is composed of experts from the field of ethics, anthropology, statistics, public health and biobanking.


07-06-2018
Genome Database of Latvian population participates in research, that is related to influence of smoked dietary products on gut microbiome composition.
Aim of the project is multidimensional investigation of the impact smoked food especially fish consumption exerts on human gut microbiome to gain insight in the responding functional pathways and metabolic consequences.
Principal investigator: Dr. biol. Ineta Kalniņa
Research is funded by the ERDF No.1.1.1.2/VIAA/1/16/128 project with the same title.
Approval by Central Medical Ethics Committee No: 1/18-06-07
26-02-2018
Genome Database of Latvian population participates in study „Development of unified therapeutic observance model for patients with inflammatory bowel disease using immunologic, molecular biology and mophological methods".

Aim of the project is to study individual differences in tiopurine metabolism and terapeutic survilance using immunological, clinical, molecular and morphological methods, in patients with inflammatory bowel disease.
Principal investigator: Dr. Polīna Zaļizko
Approval by Central Medical Ethics Committee No: 3/18-02-21
28-01-2018
Genome Database of Latvian population participates in the project „Etiology of severe influenza infection in Northern Europe (NorthernFlu)”.
Objectives and expected results
The main goal of the project is to decipher etiology of severe influenza virus (IV) infections in Northern Europe. To achieve this goal we will:
- collect NPA/S samples and data from 500 in- and 1500 out-patients with IV infections in Finland, Estonia, Norway, and Lithuania during upcoming epidemic season;
- sequence viral and genotype human genomes and perform GWA studies to identify genetic signatures associated with patient hospital admissions;
- validate GWAS results using viral and host reverse genetics in cell culture and in mouse models;
- publish the results.
Viral and host genetic factors associated with patient hospital admissions will be exploited towards development of novel approach for prediction and prevention of severe IV infections. The novel approach will allow clinicians to accurately and rapidly diagnose severe IV infections using a single patient sample. These may reduce a number of hospitalizations and healthcare costs in future.
Principal investigator: Prof. Uga Dumpis
Research is funded by the project “MOBTT39: Towards better diagnostics and treatment of severe influenza infections” (NorthernFlu), funder: European Regional Development Fund, Research Council of Estonia under Mobilitas+ programme” resources, study subject recruitment is partly supported by Genome Database of Latvian popualtion.
Approval by Central Medical Ethics Committee No: 1/18-01-24
